Isoelectric Point
iOS Universel / Education
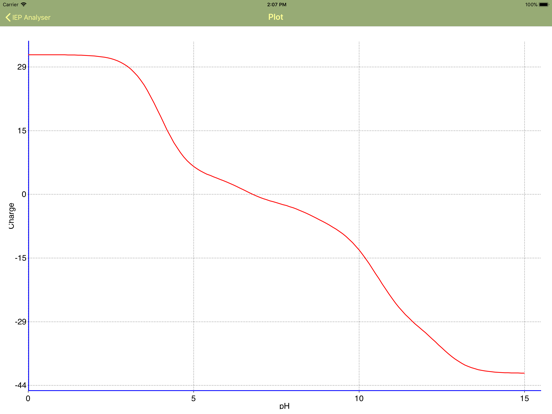
Isoelectric Point (IEP) helps to estimate the charge of macromolecule (proteins, peptides, amino acids, DNA, etc.), consisting of basic and acidic residues as a function of pH value of solution. The application graphically represents the charge fluctuation over the whole range of pH values and finds out the isoelectric point value. The application can extract the protein sequence (chain by chain) from FASTA files.
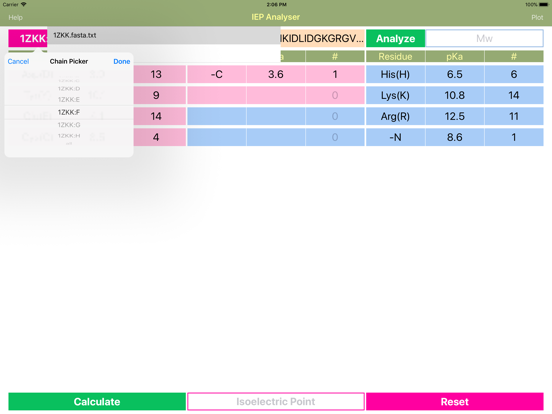
Layout of "IEP". The sequence of polymer molecule can be typed into the “Sequence” field in the single letter format (YHRK…). Alternatively, it can be uploaded from the PDB-styled FASTA file (for polypeptides) located in "Documents/IEP" folder. Pressing “Load” bar button launches file browser that lets the user to choose the FASTA file. Simple press on 'Chain' button launches chain picker dialog if multiple chains are defined in FASTA file.
If peptide is analysed, then tapping “Analyse” button, calculates the molar weight, counts and fills in the number of recognized acidic and basic residues into the appropriate cells in the amino acid info table. Cells for acidic residues are coloured red and for basic are blue. Each row provides three cells:
1. compound (amino acid) name or acronym.
2. pKa value (user defined), default values are provided.
3. Number of the particular residues in the macromolecule.
The “-C” and “-N” cells represent standard C- and N-terminus respectively. If peptide has been terminally modified, then these fields data must be changed accordingly.
The “Calculate” button initiates the analysis of the table data and outputs the IEP value. Important! Table data can be fully customized after or without any sequence analysis! IEP calculation uses table values, whatever their origin is .
Long press on 'Reset' button removes amino acid data from table if the analysed molecule is not based on amino acid sequence or alternatively brings it back if peptide is analysed.
Short Tap on reset button simply resets data in table.
Application provides data for most common acidic and basic amino acid residues, but every cell can be updated! Sequence analysis reverts the table back to its original state.
Important points:
Special attention should be paid for number of -C and -N termini and their pKa values, as they depend on functional derivaterization and number of chains being analysed.
If user updates any of the data cells, the program will not start to plot until user press calculate button and only if the update is accepted.
The cells are colored according to the data input allowed (unchangeable): red for acidic and blue for basic function. However both acidic and basic residue chemical groups are usually provided with pKa values. In case that pKb value is provided, it can be easily converted to pKa using: pKa=pKw-pKb or pKa=14-pKb (at 25°C). User should check what the primary chemical property of the residue is: the proton donor, gaining negative charge (acidic) or proton acceptor, gaining positive charge (basic).
Unrecognized amino acid residue letters are skipped, but can be added manually to the available cells.
The empty cell sets are available for custom input.
The pKa values can be changed in table to fit the desired model.
The application assumes no influence between residues.
The application assigns dot as a decimal separator.
Quoi de neuf dans la dernière version ?
Compatibility update.